Week 04 — Protein Design Part I
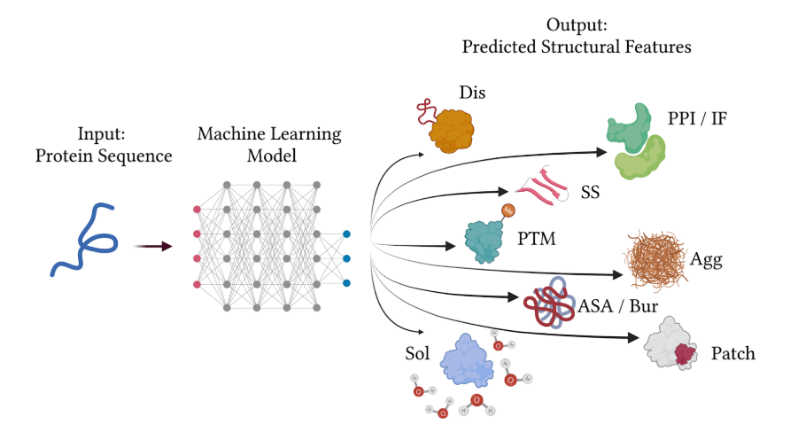
This week begins Protein Design (Part I) — modern sequence/structure tools for evaluating and designing proteins.
- This session introduces Protein Design foundations: what sequence/structure information is, how to find it, and how modern ML tools have changed design workflows. Core resources (converted from the course page) RCSB PDB — archive and tools for experimentally-determined macromolecular structures, with powerful search/visualization. UniProt / UniProtKB — comprehensive, curated protein sequence and functional knowledgebase; tightly linked with PDB and other resources. Predicted structures — both resources now surface computationally predicted structures for many proteins, complementing experimental data. (Slides and recording will be posted here when available.)
- Warning Homework details will be added here when posted on the course site. Context (converted from the course page) Use RCSB PDB to explore 3D structures and annotations of your protein of interest. Use UniProt/UniProtKB to obtain sequence and functional information and navigate cross‑links to structural resources. Note that these databases increasingly include computationally predicted structures alongside experimental entries.
- Info Recitation slides and recording will appear here when posted.
- Tip This week’s lab is entirely in‑silico — no in‑person wet‑lab. Concepts You’ll Learn Protein prediction tools Sequence recovery models Generative models Designing toward desired physicochemical properties Lab Protocol The protein design lab will be entirely in‑silico; you’ll run a variety of models to test different protein designs.
- Info No additional readings were posted on the course site at the time of conversion. This section will be updated if materials appear.